 Image credit: Oliver Mounsey
Image credit: Oliver Mounsey
 Image credit: Oliver Mounsey
Image credit: Oliver Mounsey
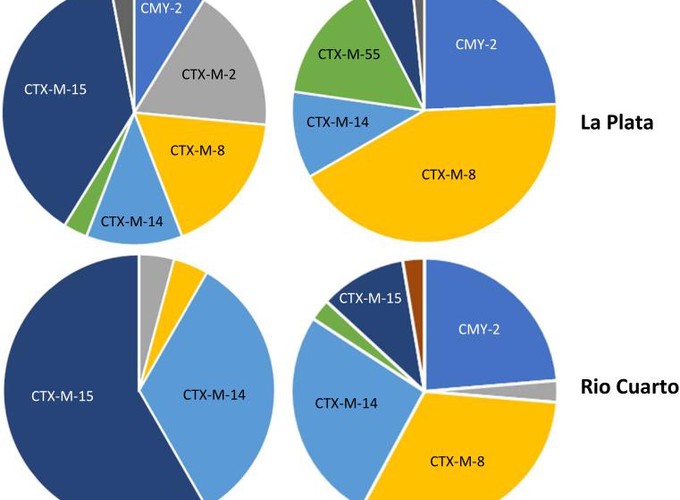
Control measures are being introduced globally to reduce the prevalence of antibiotic resistant (ABR) bacteria on farms. However, little is known about the current prevalence and molecular ecology of ABR in key opportunistic human pathogens such as Escherichia coli on South American farms. Working with 30 dairy cattle farms and 40 pig farms across two provinces in central-eastern Argentina, we report a comprehensive genomic analysis of third-generation cephalosporin resistance (3GC-R) in E. coli. 3GC-R isolates were recovered from 34.8% (cattle) and 47.8% (pigs) of samples from faecally contaminated sites. Phylogenetic analysis revealed substantial diversity suggestive of long-term horizontal transmission of 3GC-R mechanisms. Despite this, mechanisms such as CTX-M-15 and CTX-M-2 were detected more often in dairy farms, while CTX-M-8 and CMY-2, and co-carriage of amoxicillin/clavulanate resistance and florfenicol resistance were more commonly detected in pig farms. This suggests different selective pressures of antibiotic use in these two animal types, particularly the balance of fourth-versus third-generation cephalosporin use, and of amoxicillin/clavulanate and florfenicol use. We identified the β-lactamase gene blaROB in 3GC-R E. coli, which has previously only been reported in the family Pasteurellaceae, including farmed animal pathogens. blaROB was found alongside a novel florfenicol resistance gene – ydhC – also mobilised from a pig pathogen as part of a new plasmid-mediated composite transposon, which is already widely disseminated. These data set a baseline from which to measure the effects of interventions aimed at reducing on-farm ABR and provide an opportunity to investigate zoonotic transmission of resistant bacteria in this region.