 Image credit: Carlos Reding
Image credit: Carlos Reding
 Image credit: Carlos Reding
Image credit: Carlos Reding
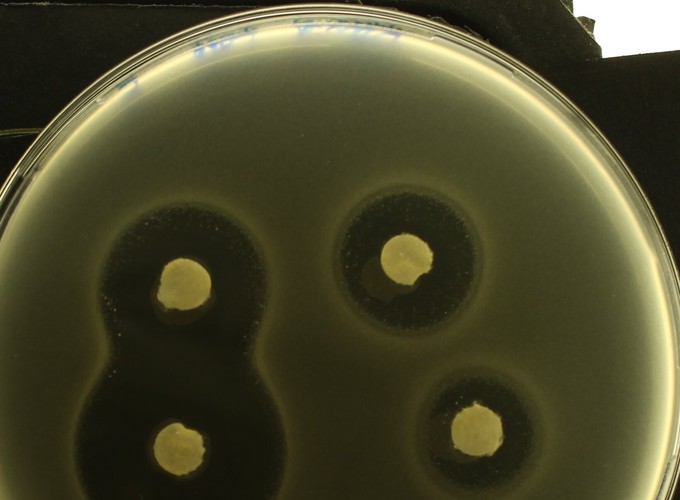
Fisher suggested advantageous genes would spread through populations as a wave so we sought genetic waves in evolving populations, as follows. By fusing a fluorescent marker to a drug efflux protein (AcrB) whose expression provides Escherichia coli with resistance to some antibiotics, we quantified the evolution and spread of drug-resistant E. coli through spacetime using image analysis and quantitative PCR. As is done in hospitals routinely, we exposed the bacterium to a gradient of antibiotic in a ‘disk diffusion’ drug susceptibility test that we videoed. The videos show complex spatio-genomic patterns redolent of, yet more complex than, Fisher’s predictions whereby a decelerating wave front of advantageous genes colonises towards the antibiotic source, forming bullseye patterns en route and leaving a wave back of bacterial sub-populations expressing acrB at decreasing levels away from the drug source. qPCR data show that E. coli sited at rapidly-adapting spatial hotspots gain 2 additional copies of acr, the operon that encodes AcrB, within 24h and imaging data show resistant sub-populations thrive most near the antibiotic source due to non-monotone relationships between inhibition due to antibiotic and distance from the source. In the spirit of Fisher, we provide an explicitly spatial nonlinear diffusion equation that exhibits these properties too. Finally, linear diffusion theory quantifies how the spatial extent of bacterial killing scales with increases in antibiotic dosage, predicting that microbes can survive chemotherapies that have been escalated to 250× the clinical dosage if the antibiotic is diffusion-limited.